XGBoost: A Scalable Tree Boosting System
Chen and Guestrin (2016)
22 Sep 2023
By the end of this talk…
You should understand every word of
XGBoost: A Scalable Tree Boosting
And why you should use XGBoost!
Let’s try it!
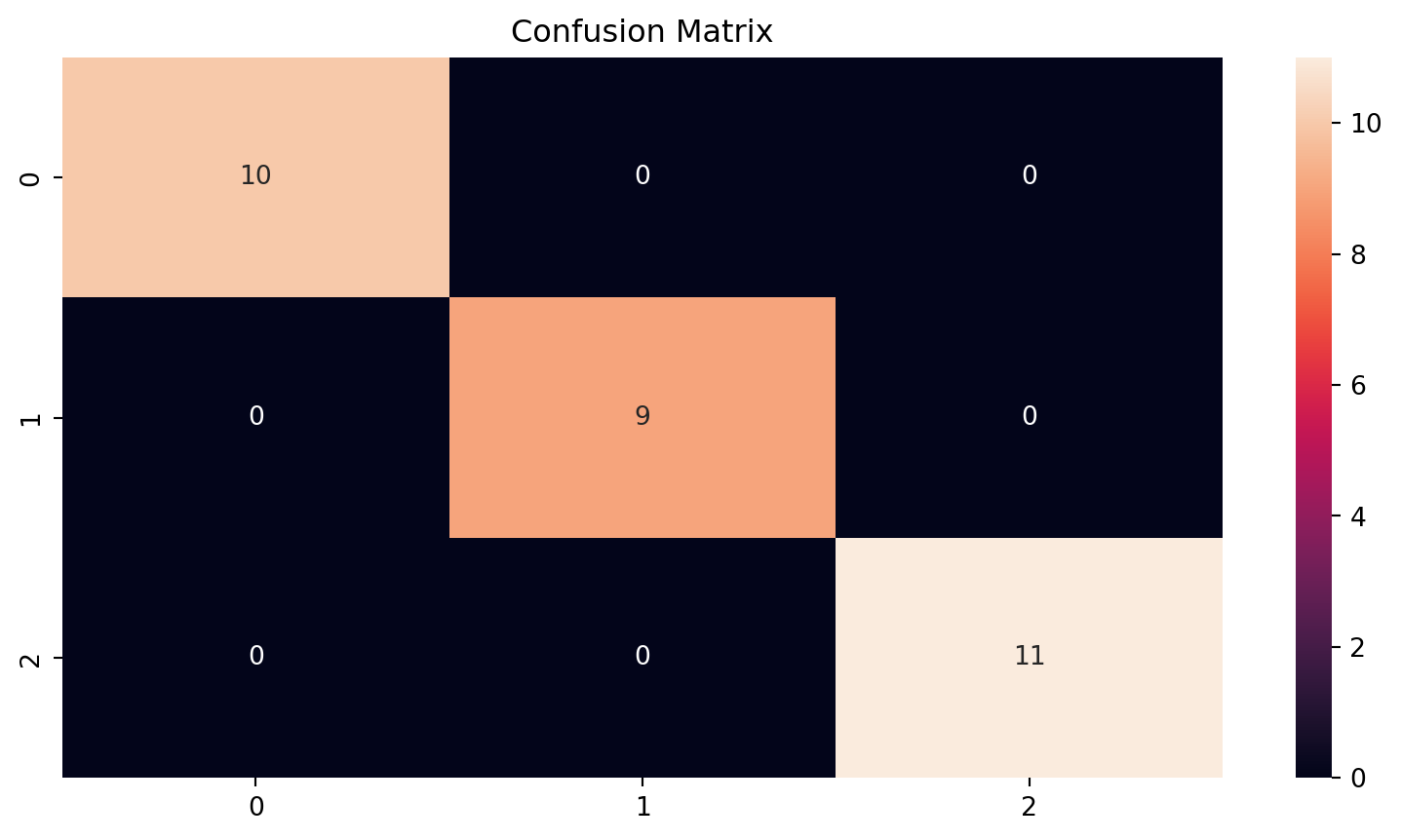
- Using the IRIS dataset from Scikit-Learn: small dataset with iris (flower) data
- We get very good classifications out of the box
from xgboost import XGBClassifier
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
data = load_iris()
X_train, X_test, y_train, y_test = train_test_split(
data['data'], data['target'],
test_size=.2, random_state=42
)
bst = XGBClassifier()
bst.fit(X_train, y_train)
preds = bst.predict(X_test)Background
From Zero to Gradient Boosting
Supervised Learning
- Supervised learning is where your training data has the desired solutions (labels).
- The model learns a function that maps inputs to correct outputs, which it can use on new, unlabled data to make accurate predictions.
Taken from 5th semester MI course at AAU
Defining an objective
- How do we measure how well the model fit the data? \[ obj(\theta)=L(\theta)+\Omega(\theta) \]
- Objective function = training loss + regularization term
- Training loss: how predictive our model is
- Regularization: helps prevent overfitting by penalizing complexity
- The goal: a simple yet predictive model
Decision Trees

Ensemble Learning
- Simple models like Decision Trees and Linear Regression are limited
- We can boost performance with ensemble learning
- Instead of creating complex model:
- Create lots of simple models (w. weak learners)
- Combine them into a meta-model
- The output is a weighted voting of the output from each simple model
Boosting
We can boost by sequentially training and combining weak learners, each trying to correct its predecessor.
- Start with a weak learner
- Calculate the errors of this initial model
- Fit another weak learner, this time focusing on the errors made by the previous model
- Combine the weak models through weighted averaging
- Repeat 2-4 until we have enough models or no further improvements can be made
Boosting
Boosting is where you learn \(F(x)\) as the sum of \(M\) weak learners. \[ F(x)=\sum^{M}_{i=1}f_{i}(x) \]
where \(f_{i}(x)\) is the weak learner.
Gradient Boosted Trees
- Same as boosting, except:
- Instead of fitting to errors from prev. trees, we fit to the negative gradients w.r.t the prediction of the prev. trees.
eXtreme Gradient Boosting
Putting it all together…
Contributions
- Scalable, end-to-end tree boosting system1
- Theoretically justified weighted quantile sketch for efficient proposal calculation2
- Sparsity-aware algorithm for parallel tree learning3
- Cache-aware block structure for out-of-core tree learning4
How XGBoost learns trees
Our model is of the form
\[ \hat{y}_{i}=\sum^{K}_{k=1}f_{k}(x_{i}),\quad f_{k}\in\mathcal{F} \]
where
- \(K\) is the number of trees,
- \(f_k\) is a function in the functional space \(\mathcal{F}=\{ f(x)=w_{q(x)} \}\), which is the set of all possible CARTs,
- \(q\) maps a sample to a leaf,
- and \(w_j\) is the weight for a leaf \(j\).
How XGBoost learns trees
The idea is we try to optimize
\[ obj(\theta)=\sum^{n}_{i}l(y_{i},\hat{y}_{i})+\sum^{K}_{k=1}\omega(f_{k}) \] where \(\omega(f_{k})\) is the complexity of the tree \(f_{k}\).
But… how do we actually learn the trees? What about their structure and leaf scores?
How XGBoost learns trees
We learn the trees additively!
\[ \begin{align} &\hat{y}^{(0)}_{i}=0 \\ &\hat{y}^{(1)}_{i}=f_{1}(x_{i})=\hat{y}_{i}^{(0)}+f_{1}(x_{i}) \\ &\hat{y}^{(2)}_{i}=f_{1}(x_{i})+f_{2}(x_{i})=\hat{y}_{i}^{(1)}+f_{2}(x_{i}) \\ &\quad\cdots \\ & \hat{y}^{(t)}_{i}=\sum^{t}_{k=1}f_{k}(x_{i})=\hat{y}_{i}^{(t-1)}+f_{t}(x_{i}) \end{align} \]
How do we know which tree we want at each step?
The one that optimizes our objective:
\[ \begin{align} obj(t) &= \sum^{n}_{i=1}l(y_{i},\hat{y}^{(t)}_{i})+\sum^{t}_{i=1}\omega(f_{i}) \\ &=\sum^{n}_{i=1}l(y_{i},\hat{y}^{(t-1)}_{i}+f_{t}(x_{i}))+\omega(f_{t})+\text{constant} \end{align} \]
However, we can’t optimize this objective function with traditional methods in Euclidean space - because our model takes functions as parameters.
And what about the regularization term?
Remember our definition of a tree: \[ f_{t}(x)=w_{q(x)},\quad w\in \mathbb{R}^{T}, q:\mathbb{R}^{d}\rightarrow \{1,2,\dots ,T\} \] \(w\) is the vector score on leaves, \(q\) is a function that assign each data point to the corresponding leaf, and \(T\) is the number of leaves.
We define the complexity as \[ \omega(f)=\gamma T+ \frac{1}{2}\lambda \sum^{T}_{j=1}w^{2}_{j} \]
Learning the tree structure
Use the Exact Greedy Algorithm:
- Start with single root - contains all training examples
- Iterate over all features and values per feature1 and evaluate each possible split gain
- Output split with best score
The gain for the best split must be positive, otherwise we stop growing the branch
Gain
We try to split a leaf into two leaves, and the score it gains is \[ Gain=\frac{1}{2}\left[ \frac{G^{2}_{L}}{H_{L}+\lambda}+ \frac{G^{2}_{R}}{H_{R}+\lambda}-\frac{(G_{L}+G_{R})^{2}}{H_{L}+H_{R}+\lambda} \right]-\gamma \]
- The score on the new left leaf
- The score on the new right leaf
- The score if we do not split
- The complexity cost by adding additional leaf
Further prevention of overfitting
XGBoost also uses two additional methods1 to prevent overfitting:
- Shrinkage: scales newly added weights by a factor \(\eta\) after each step of tree boosting.
- Column (feature) sub-sampling: randomly select a subset of features for each tree (or split), so not all features are available when finding best split. This also speeds up computations of the parallel algorithm.
Making it scalable
- Exact greedy algorithm is impossible to do efficiently when the data doesn’t entirely fit in memory - or when in a distributed setting.
- Solution: the approximate algorithm.
- It finds split points for continuous features by using histogram to approximate quantiles of the data distribution
- Don’t consider every feature value, just the boundary values as potential splits
- Uses weighted quantile sketches to determine the bin boundaries
Sparsity-aware Split Finding
- Handle sparse data by adding default directions to each tree node.
- This also exploits the sparsity to make computation complexity linear with non-missing entries in input
- This lead to 50x speedup over naive version
Block Structures for Parallel Learning
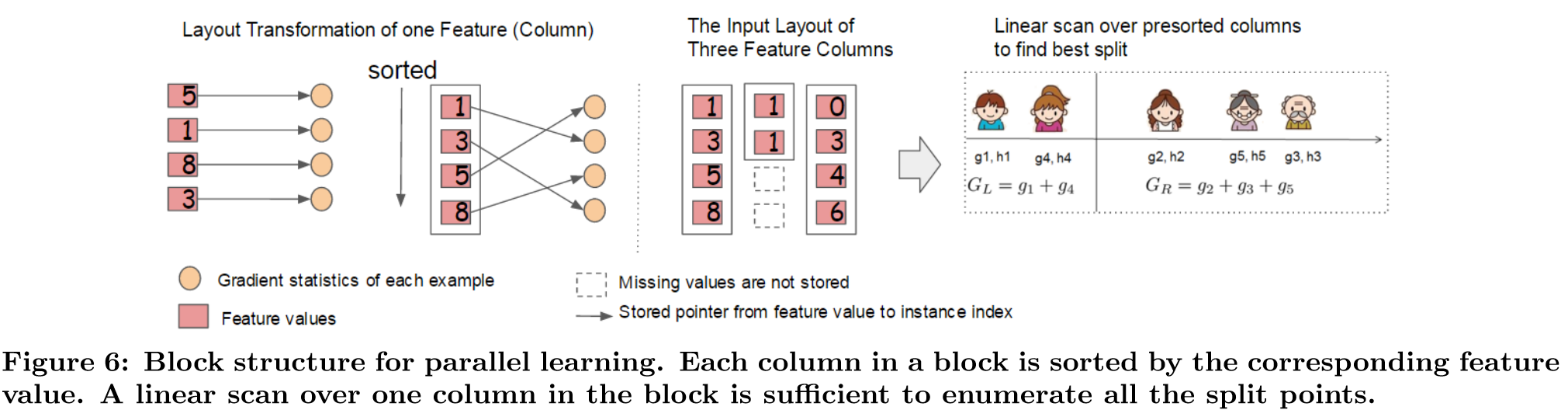
Chen and Guestrin (2016)
- Sorting the data takes most time
- Reduce cost by storing data in (in-memory) blocks
- Stored in compressed column (CSC) format - each column is sorted by corresponding feature vaue
Cache-aware access
- Block structure helps computation complexity of split finding, but the new algorithm requires non-continuous memory access
- Want to get cache hits
- Solution: Use prefetching
- Cache-aware algorithm implementation of the exact greedy algorithm runs 2x fast as naive version for large datasets
- For the approximate algorithm, the problem is solved by using a correct block size
Blocks for Out-of-core Computation
- Can make the system even more performant by better utilizing disk space to handle data that doesn’t fit in main memory
- Divide data into multiple blocks - store them on disk
- Use independent thread to pre-fetch the block into main memory buffer, so computation & disk reading happens concurrently
- Also uses block compression and block sharding
Conclusion
Two Strong Points of the Article
- True end-to-end system optimization. Very impressive focus on creating a scalable, performant system on all levels: distributed hardware, hardware, software, and algorithm.
- Impressive flexibility, enabled by accepting arbitrary loss functions: can do regression, classification, ranking, etc. with XGBoost.
Two Weak Points of the Article
- Evaluation was very focused on efficiency. Would have liked to see more about performance on various tasks.1
- The focus on so many different optimizations makes the paper very broad
Take-Home Message
- Why XGBoost?
- Superior predictions to many other algorithms, especially on tabular data
- Incredibly efficient end-to-end system
- And hopefully every word of “XGBoost: A Scalable Tree Boosting System” now makes sense